OGSync
OGSync is an Organelle Genome Synchronizer to manage a local dataset for easy-access to NCBI Organelle Database
OGSync: A CLI Organelle Genome Database Synchronizer












OGSync is NOSQL-based, freely-available and user-friendly database which provides a command-line-interface platform for bioinformatics researchers to manage a local database to synchronize and analysis multiple complete genome sequences, gene sequences and feature annotations of species. Currently, this toolkit provides the function of managing local database, synchronizing data with NCBI Organelle Genome database, and a tree viewer of gene sequences and feature records in the database. High availability of distributed file system interface, extensive data analysis of feature records from GenBank files, visually human view and json-based data interface makes OGSync a valuable data management system for studies in organelle genomics.
Motivation
After three-decade accumulation of high-throughput sequence data from various organisms, biology scientist have published over 23 thousands organelle genomes. These organelle DNAs are featured as a compact genome structure compared with nuclear genomes; thus, there are efficient molecular tools for the analysis of gene structure, genome structure, organelle function and evolution. However, an integrated organelle genome data management system, which could enable users to synchronize and analysis data in local computing system, has not previously been developed.
Architecture
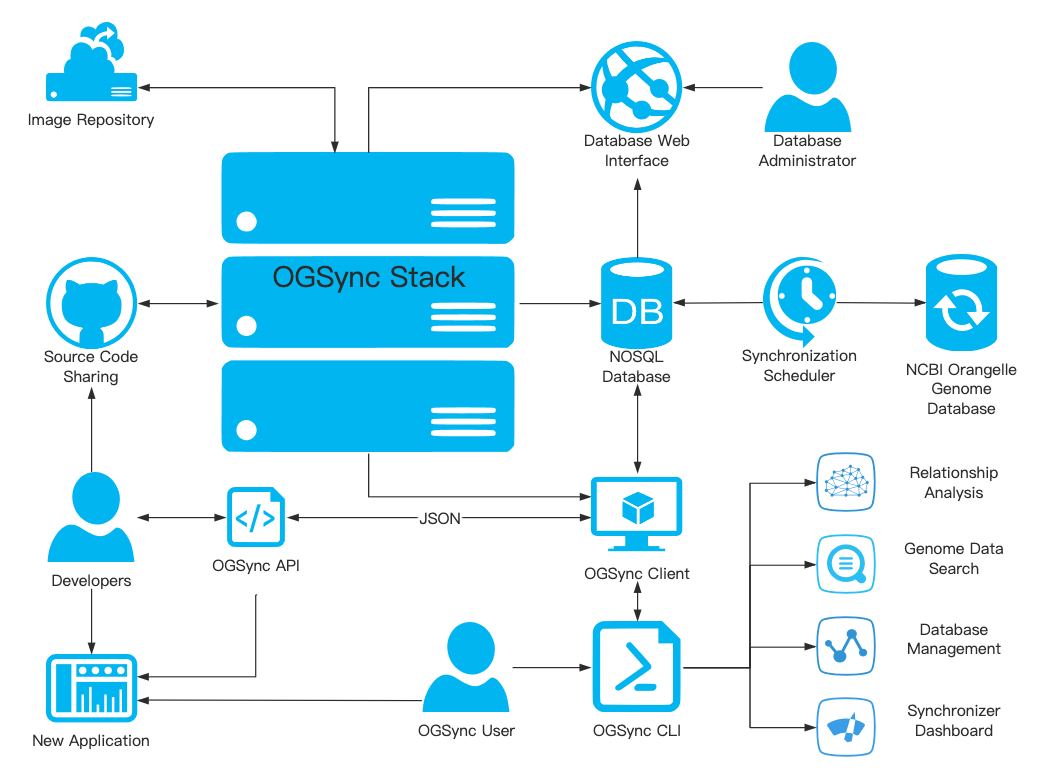
The main module of the tool is implemented by the python and can be deployed by docker engine. Users can freely build their own database and easily manage organelle genome features such as nucleotide CDS, protein, sequence, annotations and etc.
Quick Start with Docker Stack
- Ensure a docker engine is installed (see Get Docker), which supports docekr-compose utilities.
- Download the repo and unzip.
-
Start up the docker stack.
docker-compose up -d --build -
Open terminal and login OGSync container
docker exec -it ogsync /bin/bash -
Init the OGSync
OGSync init -
Update status and sync the list with NCBI database
OGSync update -
List all available refSeq, use
-rto display theremotegenome codeOGSync list -r -
Add some refSeq by using
addcommand. For example, if you want to sync the chloroplast genome of Arabidopsis thaliana to your local database.OGSync add NC_000932.1 -
Try
showcommand to display the tree view of the genomic info, or use-jargument to obtain an api-friendly response.OGSync show NC_000932.1 annotation OGSync show NC_000932.1 annotation -j -
Try more command to manage your local database and sync.
OGSync --help
Manual install without Docker Stack
-
Pull the
OGSyncimage (see OGSync image page),docker pull yiqingxu/ogsync -
Setup a mongo server and change the config file oglib/og_mongo.py in docker at
/opt/OGSync/oglibMONGO_LINK = 'mongodb://OGSync:OGSyncDocker@mongo:27017/' -
Init the OGSync, and report issue with
--debuginfo if failed.OGSync init --debug
Documentation
Please visit our OGSync Documentation page to see more.
Reference
- NCBI Organelle Genome Resources, https://ncbi.nlm.nih.gov/genome/organelle/



